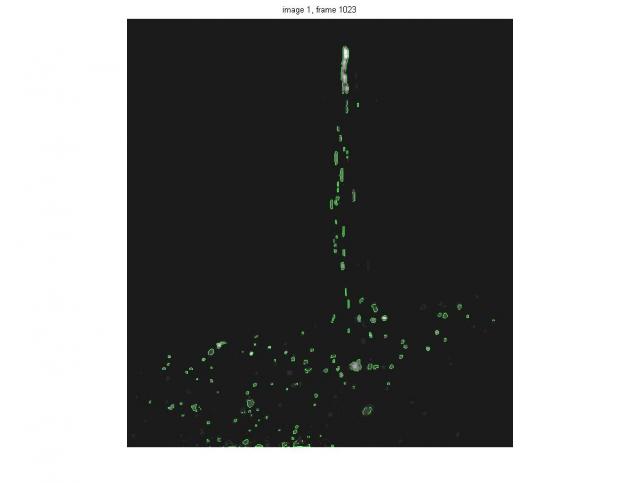
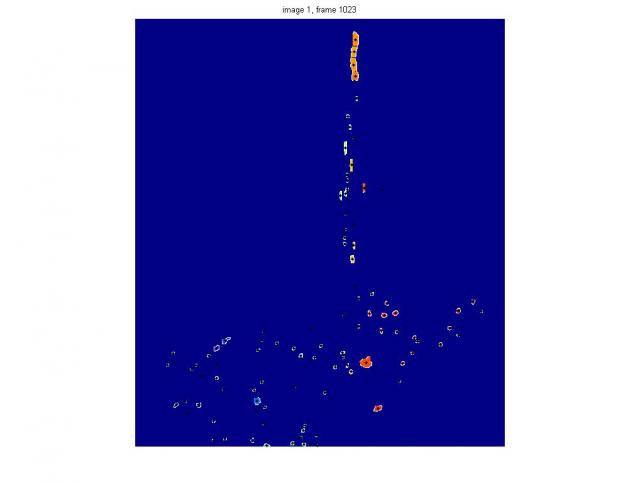
Here is the Image Processing approach to the colloid breakage identification. The test case is take from the Terradisk, Beat_Colloid folder. The main tools are very well explained on the Mathworks blog: http://blogs.mathworks.com/steve/2006/06/02/cell-segmentation/.
This is by no means the final solution, but rather a demo of the approach that I can propose. The idea is that image is segmented into 'complex shape particles' using image processing subroutines. This allows to extract the boundaries using the grey level thresholding and, in addition, to see if there are one or two (or many) 'bright islands' within that boundary.
The code is zipped in the file, segmentation_of_breakage.zip and in the subroutine colloid_image_segmentation_inpoly. These files allow for proper identification of the single-image-breakage-event, i.e. the point at which the single particle becomes two-hill particle. However, for obvious reasons, this doesn't mean that all the cameras (illumination, view angle effects, etc.) will see the "same" event at the same frame. The main problem is that even I do not know how to define properly breakage in 2D using 4 cameras. The missing step is use the output, feed it back into image-to-3D-space routine in PTV software and get 3D positions of the particles. Maybe then the discrepancies between the four camera images will be found.
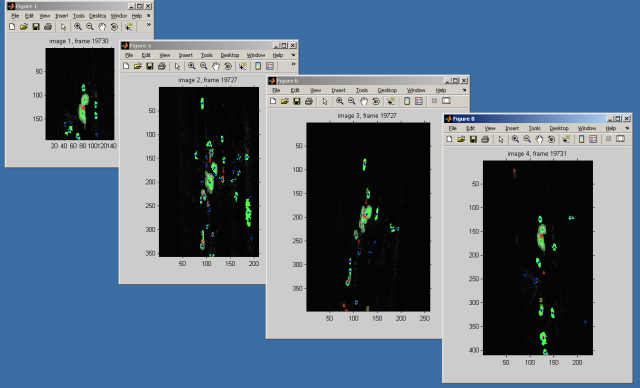
From Beat:
| Attachment | Size |
|---|---|
| segmentation_of_breakage.zip | 3.28 KB |
| screen.png | 124.04 KB |
| 2d_recognition_continued.m.txt | 2.45 KB |
Connection to 3D-PTV
The code is already given - see the branch created by Alex on SVN repository, under
\branches\extended_image_processing
The main idea of this branch is that the software can now READ the _targets files instead of creating it. Use \parameters\pft_version file, set the value to 4 (instead of 0,1,2 or 3 that are usually used, 3 is the default) and you'll get the software that READS _targets files, reorders and sorts them, and builds correspondences (stereo-matching).
Everyone can now create any type of Matlab image processing and connect it to the Matlab file write_targets.m
function write_targets(filename,stats)
xy = [stats.Centroid]';
x = xy(1:2:end);
y = xy(2:2:end);
n = [stats.Area]';
sumg = round([stats.sumg]');
nx = round([stats.MajorAxisLength]');
ny = round([stats.MinorAxisLength]');
np = length(n);
pnr = 0:np-1;
tnr = -1*ones(np,1);
fid = fopen(filename,'w');
fprintf(fid,'%d\n',np);
fprintf(fid,'%4d %9.4f %9.4f %5d %5d %5d %5d %5d\n',[pnr',x,y,n,nx,ny,sumg,tnr]');
fclose(fid);