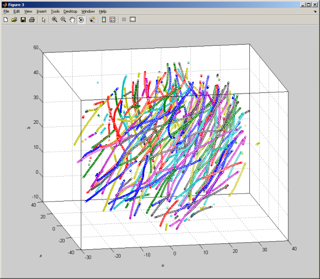
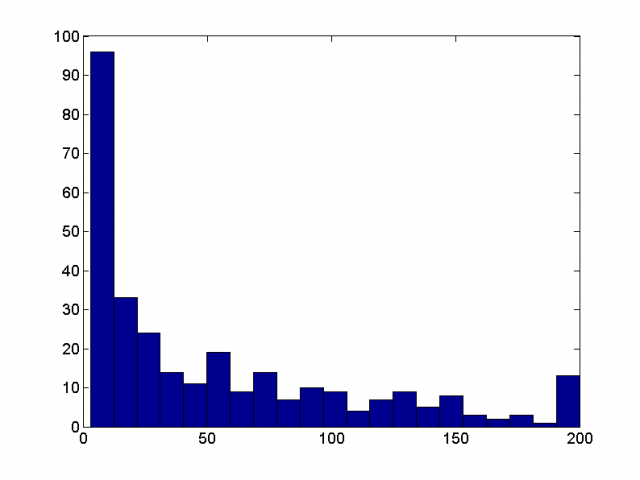
Result of our software tracking of the data posted by Greg Voth, Wesleyan University
The data is posted on http://gvoth.web.wesleyan.edu/PTV/Wes_data.htm
After some transformations (the detailed instructions will be posted soon), Alex got this:
There are 310 trajectories of various length, few longest are 200 frames (full run) long:
some e-mail that should be converted into wiki page
- The data is Wesleyan data.
- The software is 3D-PTV software, v1.02 (I installed the batteries-included-version http://ptv.origo.ethz.ch/wiki/Windows_Installer_Package)
- All the changes were performed in the /test subfolder of this installation in order to be sure that the same parameters are set up. (parameters.zip is attached).
- our coordinate system is different and it's not flexible, so I had to change it to:
- x -> z, y -> x, z -> y
- renamed image files to the format of
cam1.10001 - cam4.10200(I added 10000 for the convenience, I never sure about 0xxx format of the extension) - renamed calibration images
- av_calib1_edit -> cam1.tif
- We use 'negative' images, i.e. white dots on dark background. some matlab trick is attached
- function wesleyan_calib_img_to_zurich_img(imgfilename)
- if ~nargin
- imgfilename = 'E:\PTV\New Folder\test\cal\av_calib1_edit.tif';
- end
- cam = imread(imgfilename);
- imshow([cam,imcomplement(cam)])
- [path,name,ext] = fileparts(imgfilename);
- imwrite(imcomplement(cam),'cam4.tif','compression','none');
- created orientation files. first version was simply the copy of the information from Wes_data.htm, e.g. for camera 1:
- 650.0 70.0 650.0 <-- x,y,z camera position
- 0.2 0.0 0.0 <-- angles, 0.2 rad looking d0 1 00 0 10.0 0.0 &nownwards
- 100.0 <--- focal distance, unknown parameter
- 500.0 0.0001 500.0 <--- interface, perpendicular to the camera, radius of the tank
- the attached *.ori files are already after static (using the calibration plate) and dynamic calibration ("shaking").
- created
calblock.txtfile that numbers the points on the calibration plate from 1 to 100, such that:
- 1 ... 9
- .......
- 91 .. 100
- changed the Main Parameters to have 10 mm glass and images of the 1280 x 1024 pixels. I didn't know the pixel size, changed it to 10 micron (should not be a big problem on average) and I do not know how much it affects the final results.
- using manual tagging of points 1,9,91,100 and few iterations, the data was pretty well calibrated, due to large distance from the calibration plane, the numbers are below 1 micron accuracy.
- added dynamic calibration using first 50 frames. the parameters folder is zipped and attached.
- standard run of the 3D-PTV software (v1.02, from the standalone installation, see origo) with sequence/tracking and tracking backwards created the data which is attached in wes_data.zip. The files were converted from _targets files to the cam1,2,3,4_2D.dat using our Matlab subroutine (attached, et_targets_to_bench2D_format).
- the branch of our post_processing software (see on origo) that is called bench_3D_output creates the text file (inside the wes_data.zip) called wes_bench3d.txt. the format is as it was before: x,y,z,frameId,trajID
Hopefully, everyone can now re-run it with the same settings to get the same/close result. If possible, someone has to repeat the steps (now it's easier) and type a tutorial of how to use Wesleyan data in 3D-PTV software. Since it's a nice one-plane calibration that evolves as a dynamic calibration is added - we could consider this case as a nice tutorial for the 3D-PTV users. we can also test various dynamic calibrator algorithms on this case, as a part of the post-benchmark collaboration.
| Attachment | Size |
|---|---|
| wesleyandataprocessingandresults.zip | 998.24 KB |